uflow
U-Flow: A U-shaped Normalizing Flow for Anomaly Detection with Unsupervised Threshold.
Link to download paper:
Link to view the paper online:
This is the official code that implements the paper U-Flow: A U-shaped Normalizing Flow for Anomaly Detection with Unsupervised Threshold.

Citation
@article{Tailanian2024uflow,
title = {U-Flow: A U-Shaped Normalizing Flow for Anomaly Detection with Unsupervised Threshold},
ISSN = {1573-7683},
url = {http://dx.doi.org/10.1007/s10851-024-01193-y},
DOI = {10.1007/s10851-024-01193-y},
journal = {Journal of Mathematical Imaging and Vision},
publisher = {Springer Science and Business Media LLC},
author = {Tailanian, Mat\'ias and Pardo, \'Alvaro and Mus\'e, Pablo},
year = {2024},
month = may
}
Abstract
In this work we propose a one-class self-supervised method for anomaly segmentation in images that benefits both from a modern machine learning approach and a more classic statistical detection theory. The method consists of four phases. First, features are extracted using a multi-scale image Transformer architecture. Then, these features are fed into a U-shaped Normalizing Flow (NF) that lays the theoretical foundations for the subsequent phases. The third phase computes a pixel-level anomaly map from the NF embedding, and the last phase performs a segmentation based on the a contrario framework. This multiple hypothesis testing strategy permits the derivation of robust unsupervised detection thresholds, which are crucial in real-world applications where an operational point is needed. The segmentation results are evaluated using the Mean Intersection over Union (mIoU) metric, and for assessing the generated anomaly maps we report the area under the Receiver Operating Characteristic curve (AUROC), as well as the Area Under the Per-Region-Overlap curve (AUPRO). Extensive experimentation in various datasets shows that the proposed approach produces state-of-the-art results for all metrics and all datasets, ranking first in most MVTec-AD categories, with a mean pixel-level AUROC of 98.74%.
Code and trained models are available at https://github.com/mtailanian/uflow.

Setup
Creating virtual environment and installing dependencies
# Create conda virtual environment and activate it
conda create -n uflow python=3.11
conda activate uflow
# Install the rest of the dependencies with pip
pip install -r requirements.txt
Execution
There are three main files to execute: train.py, predict.py, and evaluate.py.
All scripts are to be run from the root directory <uflow-root>.
You might need to add this folder to the pythonpath:
export PYTHONPATH=$PYTHONPATH:<uflow-root>
Train
For training, the only command line argument required is the category:
usage: train.py [-h] -cat CATEGORY [-config CONFIG_PATH] [-data DATA] [-train_dir TRAINING_DIR]
A basic execution could be for example:
python src/train.py -cat carpet
Command line arguments:
| Argument short name | Argument long name | Description | Default value |
|---|---|---|---|
| -cat | –category | MvTec category to train. One of [carpet, grid, leather, tile, wood, bottle, cable, capsule, hazelnut, metal_nut, pill, screw, toothbrush, transistor, zipper] | None (mandatory argument) |
| -config | –config_path | Config file path. If Not specified, uses the default config in configs folder. |
None: loads the config in configs folder for the corresponding category |
| -data | –data | Folder with MvTec AD dataset. Inside this folder there must be one folder for each category. | uflow-root/data/mvtec |
| -train_dir | –training_dir | Folder to save training experiments. | uflow-root/training |
The script will generate logs inside <uflow-root>/training folder (or a different one if you changed it with the command line arguments), and will log metrics and images to tensorboard.
Tensorboard can be executed as:
cd <uflow-root>/training
tensorboard --logdir .
Predict
This script performs the inference image by image for the chosen category and displays the results.
usage: predict.py [-h] -cat CATEGORY [-data DATA]
| Argument short name | Argument long name | Description | Default value |
|---|---|---|---|
| -cat | –category | MvTec category to train. One of [carpet, grid, leather, tile, wood, bottle, cable, capsule, hazelnut, metal_nut, pill, screw, toothbrush, transistor, zipper] | carpet |
| -data | –data | Folder with MvTec AD dataset. Inside this folder there must be one folder for each category. | uflow-root/data/mvtec |
For example use like this:
python src/predict.py -cat carpet
Evaluate
This script run the inference and evaluates auroc and segmentation iou, for reproducing results.
usage: evaluate.py [-h] -cat CATEGORIES [CATEGORIES ...] [-data DATA]
[-hp HIGH_PRECISION]
| Argument short name | Argument long name | Description | Default value |
|---|---|---|---|
| -cat | –categories | MvTec categories to train. A subset of [carpet, grid, leather, tile, wood, bottle, cable, capsule, hazelnut, metal_nut, pill, screw, toothbrush, transistor, zipper]. | None: meaning to run over all categories |
| -data | –data | Folder with MvTec AD dataset. Inside this folder there must be one folder for each category. | uflow-root/data/mvtec |
| -hp | –high-precision | Whether to use high precision for computing the NFA values or not. High precision acieves slightly better performance but takes more time to execute. | False |
Example usage for two categories:
python src/evaluate.py -cat carpet grid
[Optional] Download pre-trained models - Option 1
If you are to reproduce the paper results, you could download the pre-trained models that were used to obtain the actual results, or you can even train a model with the provided code (explained in next section).
Pre-trained models for MVTec can be found in this release
[Optional] Download pre-trained models - Option 2
For downloading the pre-trained models, go to project root directory and execute the download_models.py script in the following way:
First, go to the root directory:
cd <uflow-root>
Then execute the following script:
usage: download_models.py [-h] [-cat CATEGORIES [CATEGORIES ...]] [-overwrite FORCE_OVERWRITE]
| Argument short name | Argument long name | Description | Default value |
|---|---|---|---|
| -cat | –categories | MvTec categories to download. None or a subset of [carpet, grid, leather, tile, wood, bottle, cable, capsule, hazelnut, metal_nut, pill, screw, toothbrush, transistor, zipper]. | None: meaning to download all categories at once |
| -overwrite | –force-overwrite | If a certain model is already downloaded, this flag is used to decide whether to download it again anyway or skip it. | False |
For example, to download all models for only carpet:
python download_models.py -cat carpet
For downloading models for both carpet and grid, and overwrite if already downloaded:
python download_models.py -cat carpet grid -overwrite true
To download all models at once:
python download_models.py
Troubleshooting
Sometimes, when attempting to download files too frequently, gdown gives an error similar to this:
Access denied with the following error:
Cannot retrieve the public link of the file. You may need to changethe permission to 'Anyone with the link', or have had many accesses.You may still be able to access the file from the browser:
https://drive.google.com/u/1/uc?id=12ZgoyzBWoip1FfmuEiQc0NDweXJr5uNd&export=download
In that case there are two options: wait a couple of hours (probably 24 hours), or download by hand by entering to this url.
If downloading by hand please remember to use the same folder’s structure as in Google Drive, inside the <uflow-root>/models directory.
Download data
MVTec
To download MvTec AD dataset please enter the root directory and execute the following
cd <uflow-root>/data
wget https://www.mydrive.ch/shares/38536/3830184030e49fe74747669442f0f282/download/420938113-1629952094/mvtec_anomaly_detection.tar.xz
tar -xvf mvtec_anomaly_detection.tar.xz
rm mvtec_anomaly_detection.tar.xz
If you prefer, you can also download each category independently, with the following links:
Texture categories: carpet, grid, leather, tile, wood,
Object categories bottle, cable, capsule, hazelnut, metal_nut, pill, screw, toothbrush, transistor, zipper,
Bean Tech
https://paperswithcode.com/dataset/btad
LGG MRI
https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation
ShanghaiTech Campus
https://svip-lab.github.io/dataset/campus_dataset.html
Results
Pixel AUROC over MVTec-AD Dataset
![]()
Pixel AUPRO over MVTec-AD Dataset
![]()
Segmentation results (mIoU) with threshold log(NFA)=0

## Results over other datasets


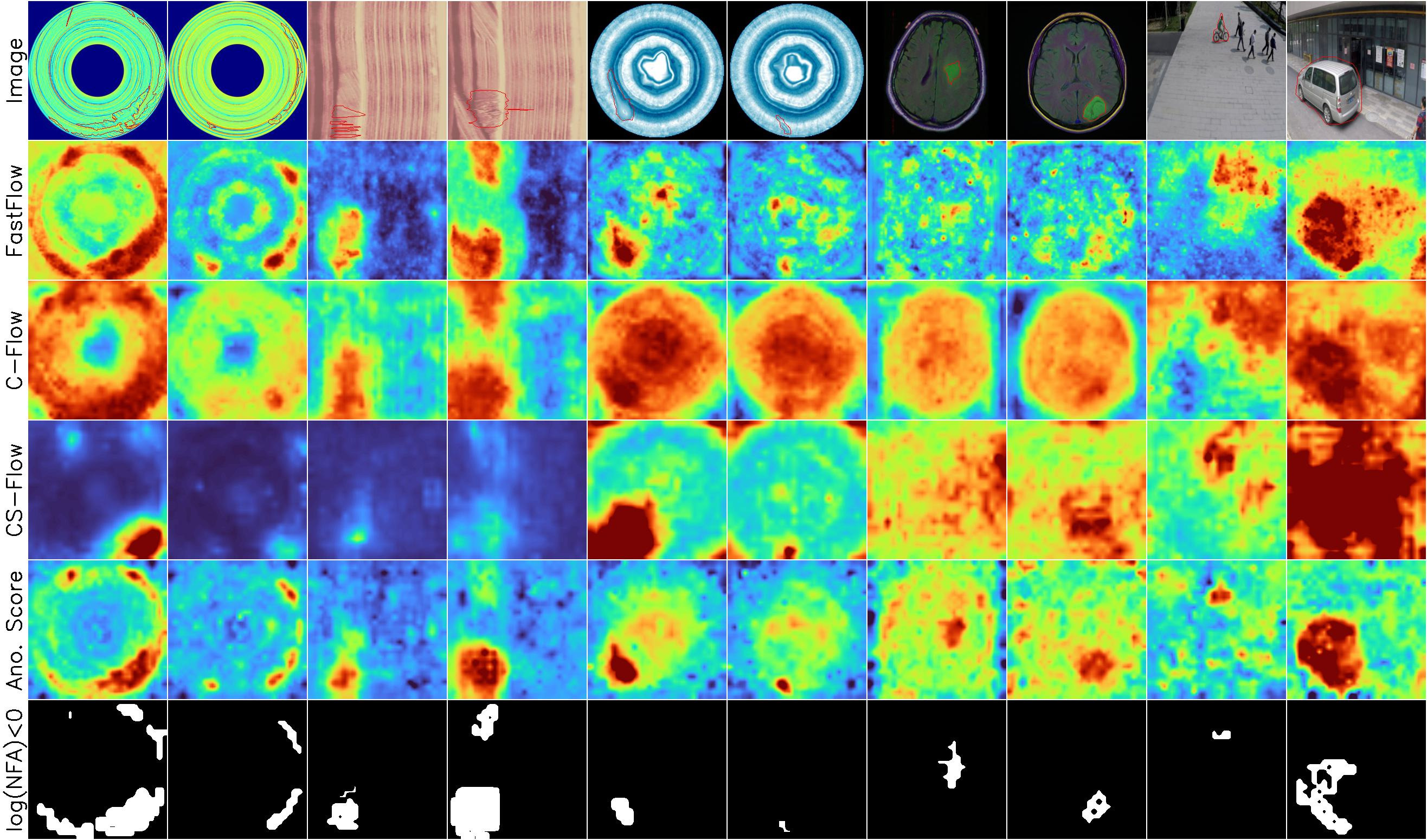
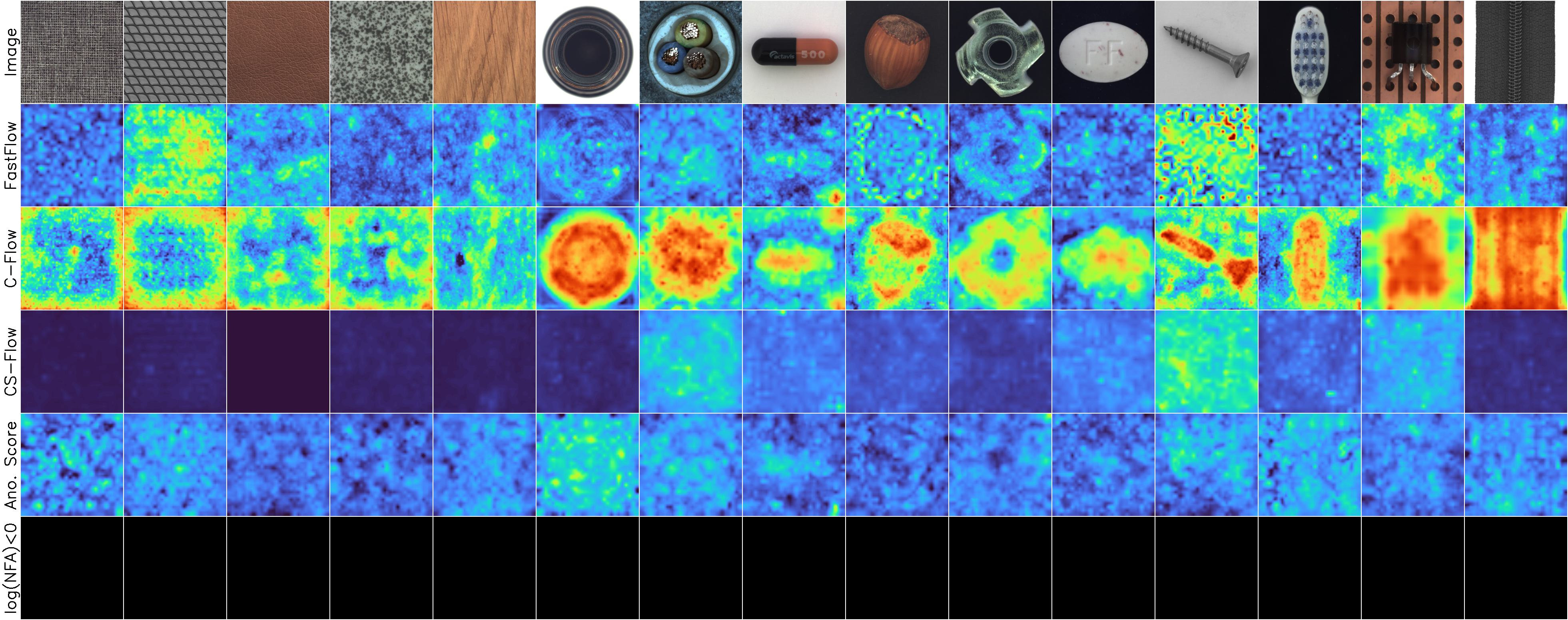
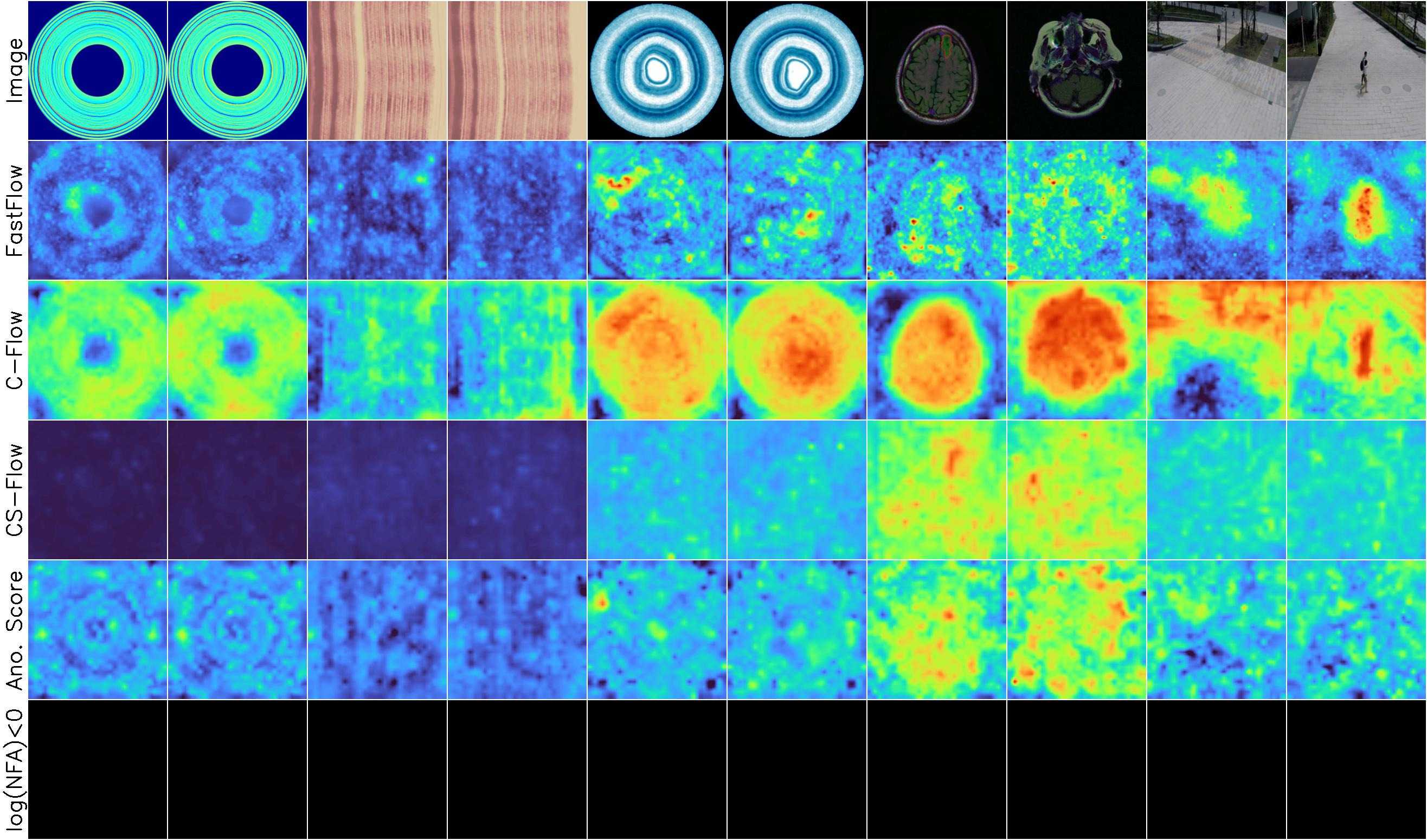
Example results
Anomalies
MVTec
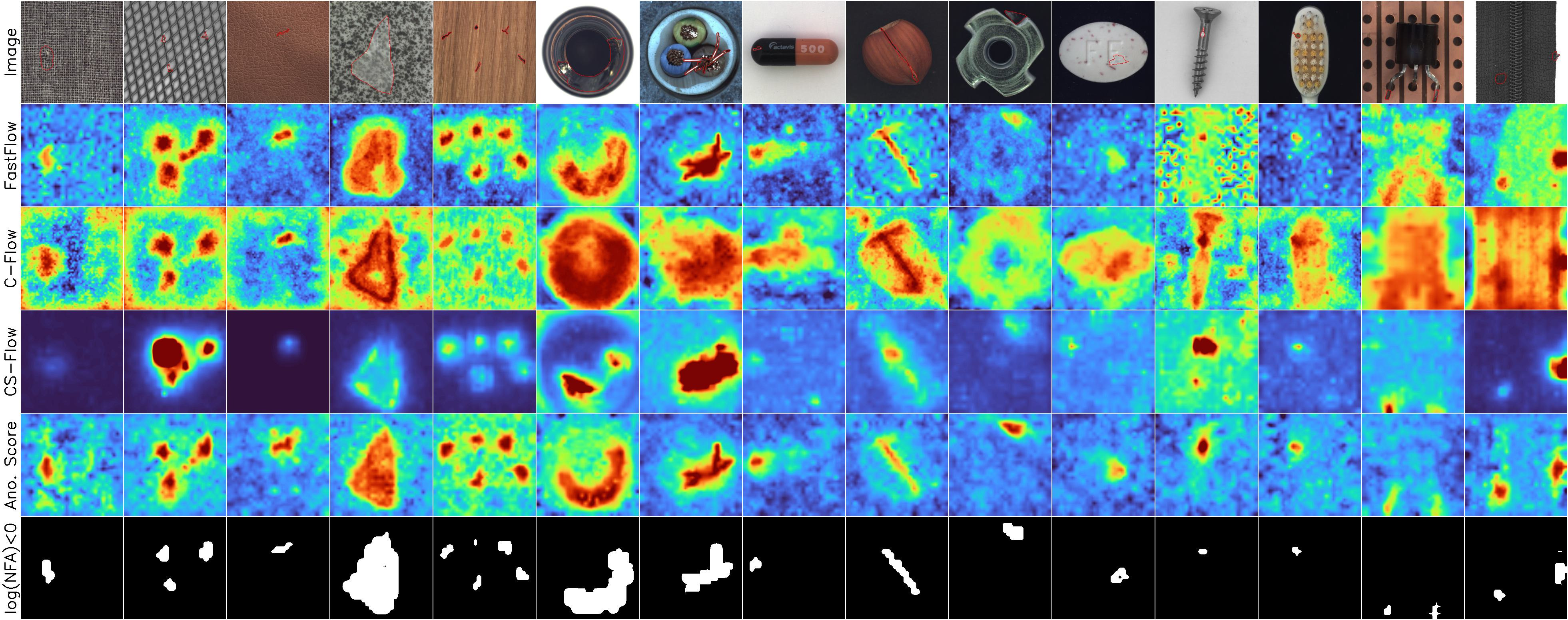
BeanTech, LGG MRI, STC
Normal images
MVTec
BeanTech, LGG MRI, STC
A note on sizes at different points
Input
- Scale 1: [3, 448, 448]
- Scale 2: [3, 224, 224]
MS-Cait outputs
- Scale 1: [768, 28, 28]
- Scale 2: [384, 14, 14]
Normalizing Flow outputs
- Scale 1: [816, 28, 28] --> 816 = 768 + 384 / 2 / 4
- Scale 2: [192, 14, 14] --> 192 = 384 / 2
/ 2 corresponds to the split, and / 4 to the invertible upsample.
Copyright and License
Copyright (c) 2021-2022 Matias Tailanian
This program is free software: you can redistribute it and/or modify it under the terms of the GNU Affero General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU Affero General Public License for more details.
You should have received a copy of the GNU Affero General Public License along with this program. If not, see http://www.gnu.org/licenses/.


